Forget silicon - SQL on DNA is the next frontier for databases

You probably don't want to see any more factoids about how much data the world generates every day. We certainly don't. Let's just agree that the data frenzy in the world today is outpacing not just our ability to comprehend numbers and qualifiers - Zettabytes, anyone? - but also our capacity to store it.
Everything is going digital, and everything is increasingly ran on applications based on algorithms trained on data, which in turn generate more data to feed more downstream applications and algorithms..you get the picture.
Simply put, at this pace, there soon won't be enough data storage and compute material to go by. Which is why people have been looking into alternative storage media for data for a while now. Using DNA to store data, strange as it may sound at first, actually makes lots of sense. And now researchers have made a breakthrough, enabling them to integrate DNA storage in PostgreSQL, a popular open source database.
DNA as an information encoding mechanism
At its core, DNA is a data storage layer. DNA is made up of four base components: Adenine, Guanine, Cytosine and Thymine (aka AGCT). From these four bases, DNA forms groups of three nucleotides (known as codons). A codon is the unit that gives our cells instructions on protein formation.
Our information technology infrastructure is based on the storage of information in bits (which are made up of two digits: 0s and 1s), whereas DNA information is stored in strings of four potential base units. To store non-genetic information in DNA, we must first translate binary data from bits to the four unit (AGCT) structure of DNA data.
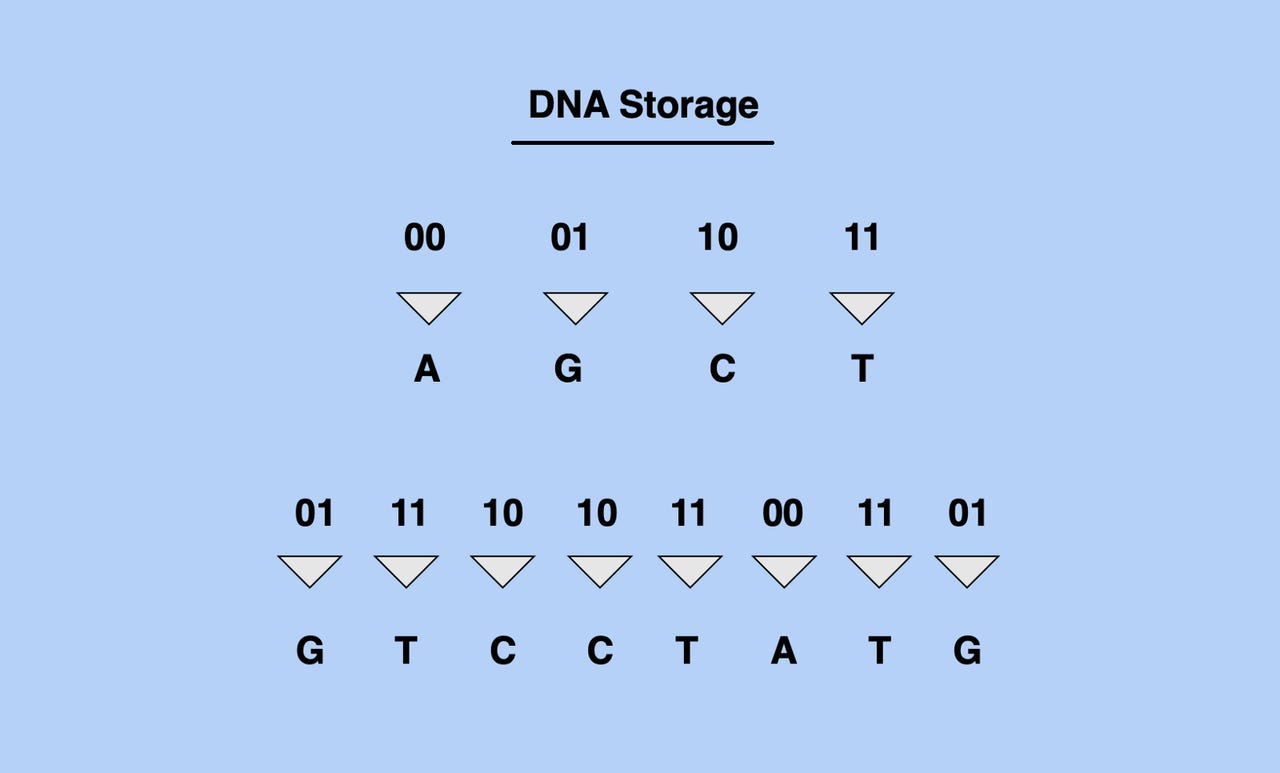
Converting Bits into DNA sequence. Image: Shaan Ray
The theoretical part is actually rather straightforward. Same as using silicon or magnetic media, which work based on their ability to store state as sequences of 1 and 0, we could use DNA, storing sequences of A,G, C, and T. But how does this work in practice - how would one go about writing and reading data in DNA?
This may sound far out, but progress in molecular technology has made it feasible - albeit not perfect. What this all means is that encoding information in a way that makes it possible to store and retrieve it on DNA actually works, leveraging DNA synthesis and sequencing, respectively.
Microsoft, for example, has showcased the world's first automated DNA data storage and retrieval system. In case you're wondering where this DNA is coming from: this is synthetic DNA, and the array that produces it is part of the system.
Naturally occurring DNA is structured in the form of a double helix with two strands of nucleotides. In contrast, DNA used for data storage is a single stranded sequence of nucleotides, also referred to as an oligonucleotide (oligo), that is synthesized using a chemical process that assembles the DNA one nucleotide at a time.
This was the starting point for our discussion with Raja Appuswamy and Thomas Heinis. Appuswamy, Assistant Professor at the Data Science Department of Eurecom, and Heinis, leader of the SCALE Lab at Imperial College London, recently published groundbreaking work on DNA storage.
Using DNA to store data in the real world
Heinis and Appuswamy published a research paper titled "OligoArchive: Using DNA in the DBMS storage hierarchy" [PDF] in the Conference on Innovative Data Systems Research. While they are not the first ones to store and retrieve data using DNA, they are the first to do this for structured data, integrating with an off-the-shelf database, and to go beyond storage, implementing compute, too.
The first thing to realize about DNA as a data storage layer is that every time a write operation is performed, an oligo has to be synthesized. How will that work in practice? Will a lab technician have to be on standby to perform this, and "refill the tank" of raw materials used for the chemical process?
Not really, and this is the value in what Microsoft has demonstrated with its automated DNA storage and retrieval system, according to Appuswamy and Heinis. What this goes to show is that it is possible to operate such an array without a human in the loop. Just like there are no humans overseeing data center daily operation, except for maintenance, the same would apply for DNA-based data centers.
Still, we are far off from replacing hard disks with synthetic DNA arrays. To begin with, contemporary technology for storing data this way is painfully slow. Initially, storing a megabyte of data took scientists a week.
Appuswamy and Heinis concurred that more work is needed on this. Although it's beyond the scope of their own research, so they can only wait for biochemical composition process to catch up, they did offer some consolation.
First, they noted that storage speed has been getting better, currently going at a few KBs/sec. Even though that's still painfully slow compared to SSDs, for example, it's quite an improvement. It may be actually acceptable for the type of use Appuswamy and Heinis see for their research, which is archive storage.
Database engines use a three-tier storage hierarchy that consists of devices with widely varying price/performance characteristics. The performance tier stores data accessed by high-performance OLTP and real-time analytics applications.
The capacity tier stores data accessed by latency-insensitive batch analytics applications. The archival tier is used to store data that is accessed very rarely, for example, during security compliance checks, or legal audits. Today tapes are typically used in this tier.
OligoArchive alters the database storage hierarchy by replacing the tape-based archival tier with a DNA-based one. Synthetic DNA is stored using extra precaution, and it's questionable how well having DNA-based storage for the average device would work. But data and databases are going to the cloud anyway, and as long as your data is safely stored in a data center, it's all a black box to end users.
Running SQL on DNA
Appuswamy and Heinis also noted that even though it's still slow, DNA storage offers great potential for parallel processing. This is because it's abundant, and cheap -- or, to be more precise, the hope is that it eventually will be. At current rates, storing a single minute of high-quality stereo would cost something like $100,000.
While it's still prohibitively expensive to use synthetic DNA for storage at scale, Appuswamy and Heinis said they expect to see cost drop in the way that is typical for every scientific and technological breakthrough, including storage technology.
If synthesizing oligos becomes financially feasible, having a lot of them to go by would be a reasonable expectation. This would mean great potential for having many DNA storage units operate in parallel. While not every aspect of every algorithm is parallelizable, for the ones that are, great speedup can be achieved. This brings us to a key point.
Until today, DNA has been used to store unstructured files, be it text or video or whatnot. What Appuswamy and Heinis have done is to integrate DNA storage in a relational database. They have taken data and queries contained in TPC-H, a standard database benchmark, and they ran TPC-H on a PostgreSQL instance. Not serial access, but selecting data at will.
Storing structured data in a database system using DNA in the back-end, and querying via SQL, is a reality today. Image: Appuswamy et.al.
The researchers built archiving and recovery tools (pg_oligo_dump and pg_oligo_restore) for PostgreSQL that perform schema-aware encoding and decoding of relational data on DNA, and used these tools to archive a 12KB TPC-H database to DNA, perform in-vitro computation, and restore it back again.
This is huge. It means that now DNA storage can also support SQL operations to selectively access and process parts of the data. Note that data is not fetched to the database for operations to be performed there. Appuswamy and Heinis found a way to do things like SQL joins in the oligos. This goes beyond biochemical storage - it also entails biochemical compute.
To do this, however, the researchers had to deal with an array of issues related to deficiencies in the technology used to encode and decode information to and from DNA. Performing operations on DNA requires specialized encoding techniques that can generate oligos suitable for biochemical manipulation. Reading DNA data is currently very error-prone, and previous efforts relied on data over-representation: data was written in many copies, so that if the original one was corrupted, backups would exist.
By contrast, Appuswamy and Heinis rely on metadata. They add some extra bits of data in the blocks they write, leveraging database schema awareness. They show this can improve density during the encoding (write) process, and assist in identifying errors during the decoding (read) process. They noted this worked better than expected -- a bit of metadata can go a long way.
Is DNA the future of data?
Even though parts of the technology stack are immature, this is a major breakthrough. Having abundant storage for data centers already is a game changer. But the implications of making something as abundant as DNA a feasible medium for storage and compute may extend beyond our wildest imagination.
This may be just the first step in this direction, but every journey begins with a first step. Appuswamy and Heinis did not work on this alone, and they won't work alone in trying to develop it further, too. Their project, OLIGOARCHIVE, has been progressing in collaboration with other researchers from Université Côte d'Azur (UCA) and CNRS were part of this, and they will be able to grow the team and expand their research.
Eurecom, CNRS, ICL, UCA, plus Helixworks, a DNA synthesis start-up, have secured EU funding to further pursue research on DNA storage. The system will be designed to support the full automated cycle of encoding data, synthesizing it as DNA and reading it back through sequencing. It will store a variety of different data types, and enable near-data processing in the storage and precise retrieval of data.
Further research into storing data in DNA will be funded by the EU
The project is funded via the Future and Emerging Technology (FET) EU initiative, which funds projects on new ideas for radically new future technologies, at an early stage when there are few researchers working on a project topic. While this seems like a perfect match, we wondered whether the researchers have been approached by commercial entities, too.
Appuswamy and Heinis mentioned that so far interest has been mainly by other researchers - with one notable exception -- Microsoft. Not that there's anything tangible in that respect, but it seems Microsoft is showing more interest than anyone else at this point.
Getting a leg up in this technology could mean owning the future, as breakthroughs in this area will have massive fallout. And this, Appuswamy and Heinis note, shows in people's attitudes:
"A couple of years back, people would dismiss this as too far out. Today, when we tell them what we are working on, they go -- tell us more".
It looks like there will be a whole lot more of those "tell us more" prompts in the future.